
Resolving Genomic Complexity with a Superior Reference
The Benthic Advantage:
A Multi-tool Platform
Our platform's performance originates from our proprietary pan-immune reference graph and our ability to leverage that graph for both assembly and variant calling, representing a significant improvement over widely available workflows.

Unprecedented Diversity and Detail
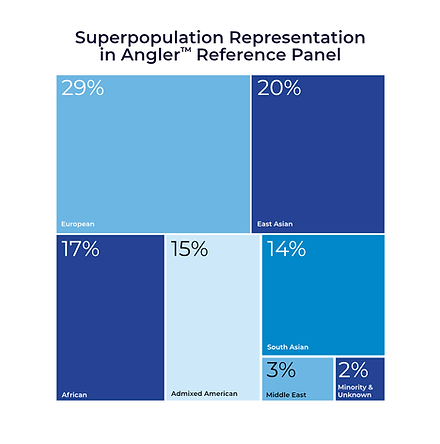
Our graph-based pan-immune reference was developed using thousands of short-read samples and incorporating several hundred high-resolution, long-read phased haplotypes of the MHC, NKC, and LRC immune regions. This provides a more comprehensive and higher-quality reference than other available resources.

Advanced Graph-Based Algorithms
Our algorithms for assembly, alignment, imputation, and phasing are specifically designed to navigate the complexity captured in our reference graph – delivering superior analytics and highly accurate allele calling and interpretation.
A Continually Improving Platform
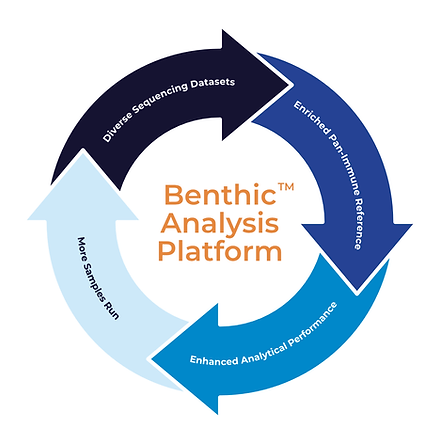
Our reference is not static. It is regularly enriched by the integration of new long-read and short-read samples. This systematic expansion improves the accuracy and comprehensiveness of subsequent results, ensuring your analysis uses the most relevant and advanced reference available for your population of interest.
The Result: Superior Performance Where It Matters Most
Our unique technological foundation translates directly into superior performance across all major genotyping platforms, including microarray, short-read NGS, and long-read sequencing.
Identify More True Variants
Confidently call more variants than other imputation panels, especially in the polymorphic "dark regions" of the genome. Angler maintains high precision while typically identifying ~50% more variants per sample than other imputation panels.


Let’s Resolve Your Most Complex Genomic Regions
Get in touch to try our platform on your own microarray or NGS data.


